Research
Human specific bacterial pathogens
Human-adapted bacterial pathogens are a specialised subset of microbes which can colonise and inflict disease on their host. These bacteria have established long evolutionary relationships with humans, which have led to the development of sophisticated mechanisms to interact with host cells, proliferate, and subvert immune responses.
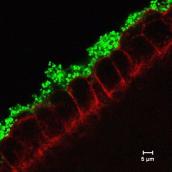
The group seeks to understand how human-specific pathogens adapt to microenvironments in the host during infection by studying their ability to colonise specific niches in the body, and evade elimination by the immune system. We use a wide range of approaches to study distantly related pathogens, Neisseria and Shigella, that invade human tissues by different routes but cause significant mortality worldwide. Furthermore, we aim to translate of our discoveries on host:pathogen interactions to the development of vaccines and therapeutics.
N. meningitidis causes bacterial meningitis and sepsis, while the gonococcus is a leading cause of genital tract infection. The group focusses on how these important pathogens and related commensal species interact with each other and their human host at the mucosal surface, and subvert the immune system. Work on fundamental steps in pathogenesis has led to the design of approaches to prevent disease through vaccination, which are being developed towards clinical trials.
We have demonstrated that oxygen acts as a crucial signal for Shigella in the intestine. Oxygen present at the epithelial surface activates Shigella genes required for host cell invasion. We have also defined a role for Shigella rhomboid intramembrane proteases. The function of rhomboids in prokaryotes has been enigmatic despite their discovery nearly 20 years ago; rhomboids selectively cleave non-functional components of respiratory complexes involved in anaerobic respiration.
A major recent focus of the laboratory is plasmid maintenance and transfer, given the importance of plasmids in the evolution of virulence and spread of antimicrobial resistance (AMR). We have examined the activity and role of toxin:antitoxin (TA) systems in Shigella, EPEC and N. gonorrhoeae, and have identified single nucleotide polymorphisms that dramatically affect their function. We are modifying TA systems for vaccine design/delivery.
Collaborators
The group has many collaborations with groups in Oxford (Martin Maiden, Department of Zoology; Susan Lea, Dunn School; Stephan Uphoff, Department of Biocehmistry), in the United Kingdom (Jun Wheeler and Ian Feavers, NIBSC; Professor Jeremy Derrick, University of Manchester), and abroad (Nathalie Balaban, HIJU, Israel; Ditlev Brodersen, Aarhus University, Denmark; Duy Pham Thanh, OUCRU, Vietnam; Philippe Sansonetti, Paris; Eduard Sanders, KEMRI:Wellcome Trust, Kenya; Ann Jerse, USUHS, USA; Eileen Barry, UMAB, USA).
Funding
Funding for the laboratory is through grants from the Wellcome Trust (Senior Investigaor Award to CMT, Collaborative award for the Gonococcal Vaccine Initiative), BBSRC, SIIPL, and the National Insitutes of Health, USA.
Members of the group have been funded by studentships (https://www.path.ox.ac.uk/content/studentship-competition-now-closed-app..., applications are open!) and external fellowships (e.g. EMBO Long and Short term fellowships, and Marie Curie Awards).
Please contact Chris to discuss.